HKBU biologist co-led team which invented a novel computational tool for cell analysis
A joint research team from Hong Kong Baptist University (HKBU), the City University of Hong Kong (CityU) and Peking University has developed a computational tool which visualises 3D shapes and changes of live cells over embryo development. This tool will revolutionise the way biologists analyse image data, furthering cell biology studies by speeding up analysis which used to take hundreds of hours by hand to a few hours by computer. Professor Zhongying Zhao of HKBU’s Department of Biology co-led this project, whose findings have been published in the journal Nature Communications titled “Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation.”
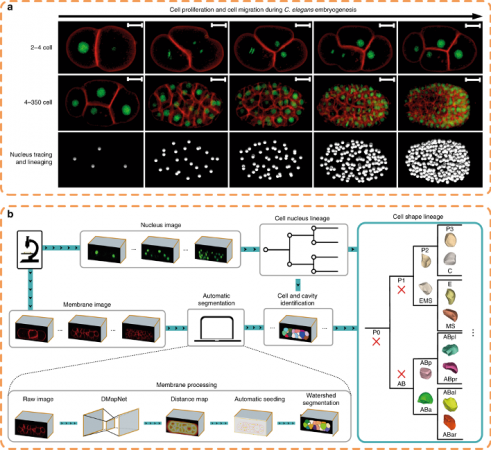
The research took advantage of the invariant development and transparent body of the nematode Caenorhabditis elegans (C. elegans). Despite previous extensive studies of cell division, cell migration and cell fate differentiation, cell morphology change during development had not yet been systematically characterised in any metazoan, including C. elegans. This knowledge gap substantially hampered many studies in both developmental and cell biology.
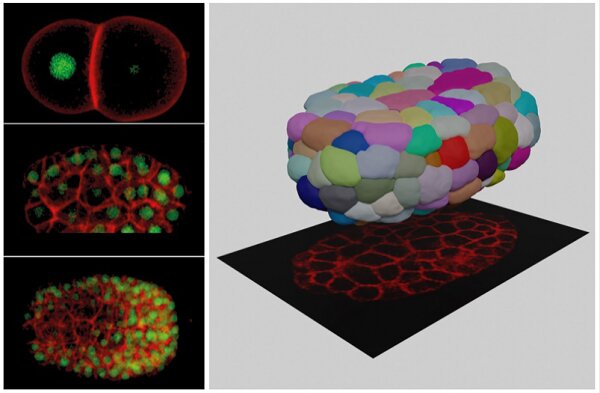
The invented tool “CShaper,” an automatic pipeline, combines automated segmentation of fluorescently labelled membranes with automated cell lineage tracing. The team applied this pipeline to quantify morphological parameters of densely-packed cells in 17 developing C. elegans embryos. Consequently, a time-lapse 3D atlas of cell morphology was generated for the C. elegans embryo from the 4- to 350-cell stages, including cell shape, volume, surface area, migration, nucleus position and cell-cell contact along with resolved cell identities.
It is anticipated that CShaper and the morphological atlas will stimulate and enhance further studies in the fields of developmental biology, cell biology and biomechanics. This opens a new avenue to study cell cycle in a live tissue or organ, including investigating whether changes in cell shape and volume will be predictive for cancerous tumour growth or stem cell differentiation.
Contact Our Researchers
DEPARTMENT OF BIOLOGY